Our lab uses a wide range of cross-scale approaches to study stem cell biology in tissues. We routinely work with 3D organoid cultures and various molecular biology techniques, including genome-engineering, advanced light and electron microscopy, and image analysis.
Intestine’s cultured in a dish
In the rapidly renewing intestinal epithelium, as stem cells differentiate, new cell contexts are formed, and cell biology such as intercellular signaling undergoes changes. Research on epithelial tissue renewal traditionally relied on snapshot observations of collected tissue states. Instead, organotypic intestines (organoids) cultured in a dish allow to investigate cell biological mechanism during dynamic stem cell fate decisions in a robust, controllable, and standardized setup.
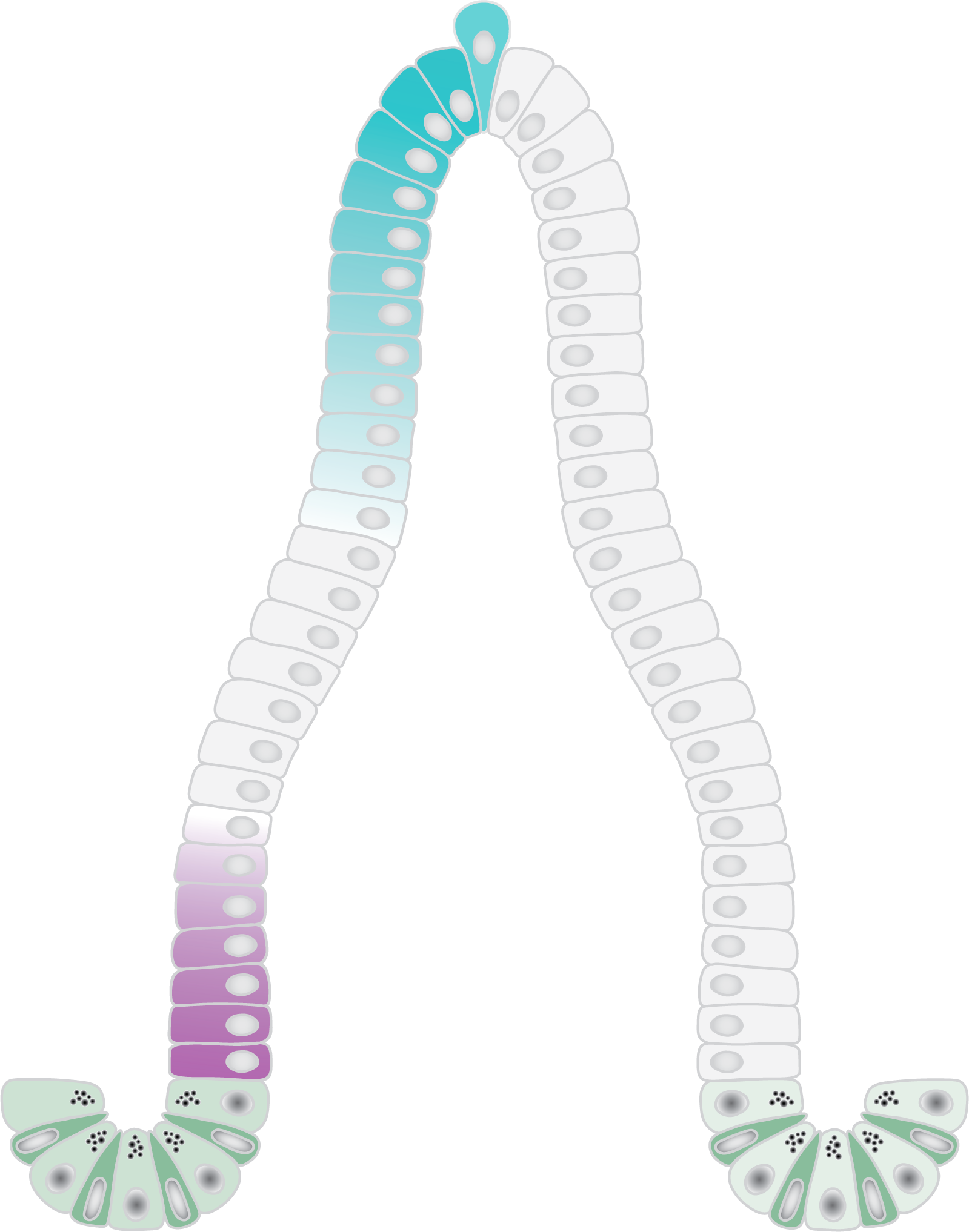
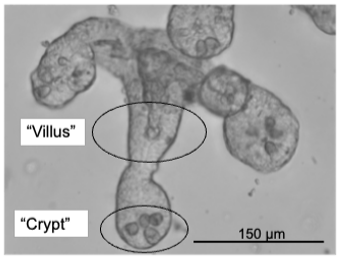
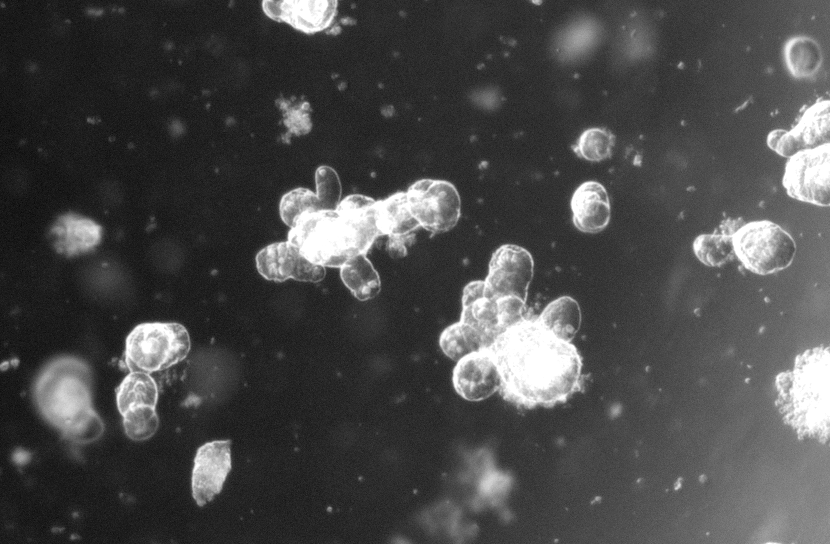
Image Description
Left: Schematic of the small intestinal epithelium.
Right: Examples of small intestinal organoids imaged in a single Z-plane (top image) or in multiple planes to cover the entire organoid (bottom image).
To grow intestinal organoids, we recreate the intestinal microenvironment in a dish. The forming organoids have the remarkable ability to self-organize into 3D structures that closely resemble the architecture, epithelial cell composition and signaling patterns of the in vivo intestinal epithelium. Stem cell-mediated tissue renewal is faithfully recapitulated and can be scored by assessing organoid growth. We apply to the organoids genetic-or drug manipulations and perform high-resolution live-cell imaging. This allows us to ultimately investigate cell biology dynamics of stem cells at single-cell resolution, yet within their 3D epithelial environment.
Volume Electron Microscopy
Video Description
With SBF-SEM, sequential intestinal tissue slices are automatically cut by an ultramicrotome within the SEM chamber and each newly exposed sample face is imaged, generating high resolution, 3D reconstructed images.
Tissue functioning of multicellular-tissues in our body relies on continuous communication between tissue cells. While the identity of the signals is intensively investigated, the arrangement of the endomembrane organelles through which the signals must traverse to be released for multi-cellular cross talk remain largely unknown. Our lab uses Volume Electron Microscopy (VEM), which provides the unique path to resolve the ultrastructure of the cellular organelles in a full tissue context. Specifically, we use Serial Block Face Scanning Electron Microscopy (SBF-SEM) as a VEM technique, which combines in-situ tissue sectioning with the high-resolution imaging of a scanning electron microscope, enabling the collection of serial images for 3D reconstruction of large tissue volumes. Using SBF-SEM our lab systematically resolves, and subsequently segments and models, spatiotemporal endomembrane organelle arrangements in a multi-cellular tissue context depend manner.
Image analysis
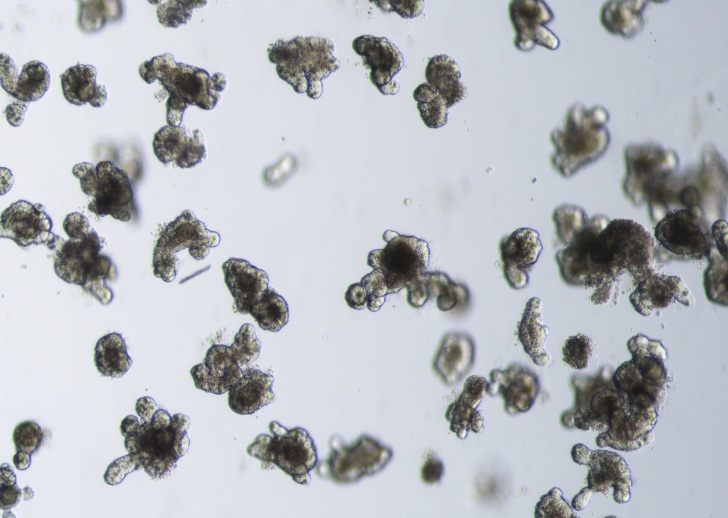

In the lab we routinely apply and develop automated image analysis tools to assess intestinal stem cell functionality within imaged organoids. We set-up imaging analysis tools to follow the growth of the organoids and extract key features such as number and width of crypt domains, as well as organoid lumen area.
Image Description
Automated intestinal organoid image analysis includes image preprocessing, reformatting and unification. We train networks for segmentation, allowing for identifying and separating different objects within the images and extract quantitative data.